Human Cell Reprogramming
iPS cell technology is a powerful tool in the field of biomedical science, and has become indispensable for drug screening in particular. Our company’s services are designed to provide researchers with the iPS cells they need for their work. Furthermore, by applying genome editing technology to both these original iPS cells and standard iPS cells, we can also prepare iPS cells that can be used as future disease models.
How to establish human iPS cells
This service provides iPS cells generated by introducing an episomal vector into peripheral blood mononuclear cells (PBMCs).
Contract generation of human iPS cell lines by genome-editting
-
1. PBMC Preparation (1 week)
Please provide PBMCs, which can be supplied by the client or purchased from FUJIFILM Wako . We will culture the cells from a frozen stock and use them for the iPS cell generation process.
*Note: The PBMCs must have a cell count of 1.0 x 10^7 cells or more at the time of freezing, be anonymous, and be negative for HIV, HBV, and HCV. -
2. Gene Transfer and Cell Culture (8-16 weeks)
We will introduce an episomal vector into the prepared PBMCs to establish iPS cells. StemFit AK02N will be used as the culture medium. -
3. Freezing iPS Cells
When the iPS cells have proliferated to an appropriate level, they are gently isolated using a medium supplemented with Y-27632 to prevent cellular damage.
They are prepared to a concentration of 1−5×105 cells/vial and cryopreserved using STEM-CELLBANKER. -
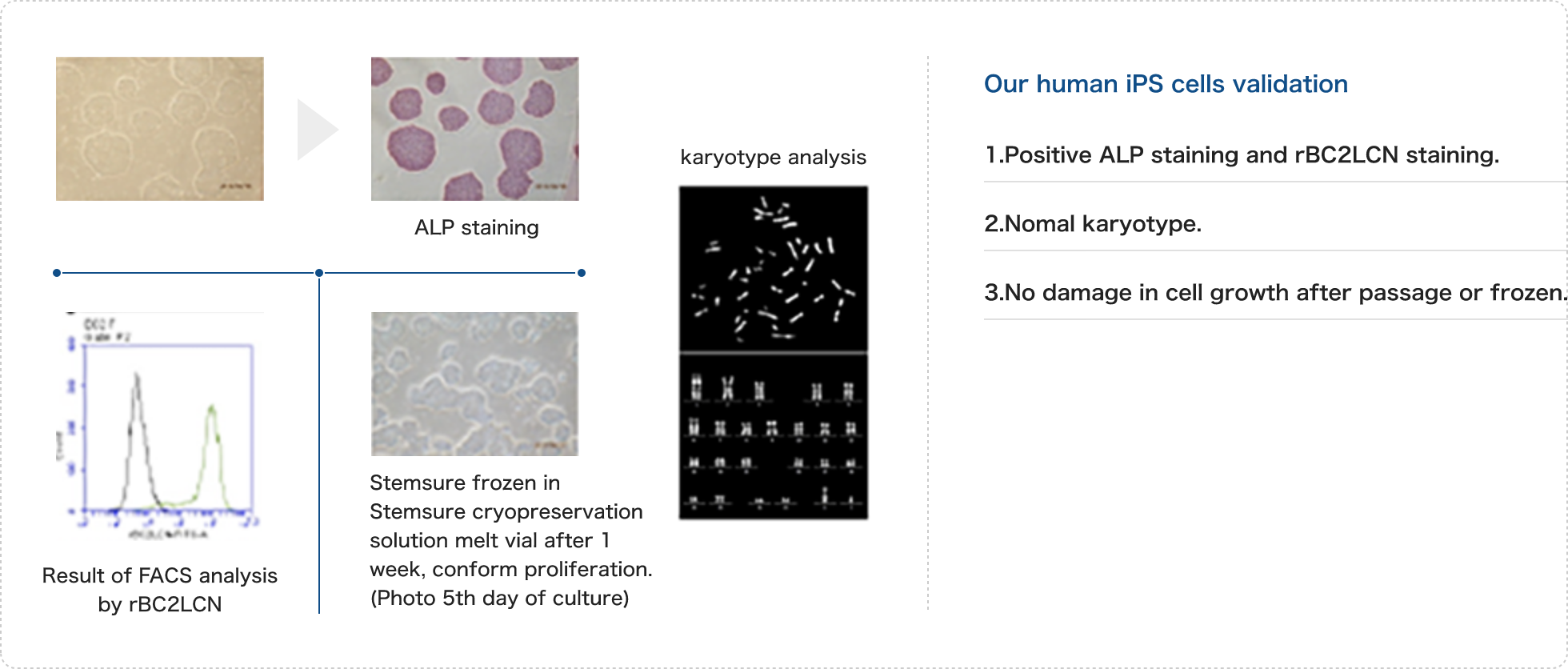
Quality Control(1-3 weeks)
iPS cells are known to exhibit various characteristics from one clone to another. Therefore, our company performs an analysis on a selection of iPS cell-specific properties to confirm the quality of the established cell lines (some analyses are optional).- iPS Cell Confirmation: Staining of iPS Cell-Specific Lectin (Live Cell Staining)
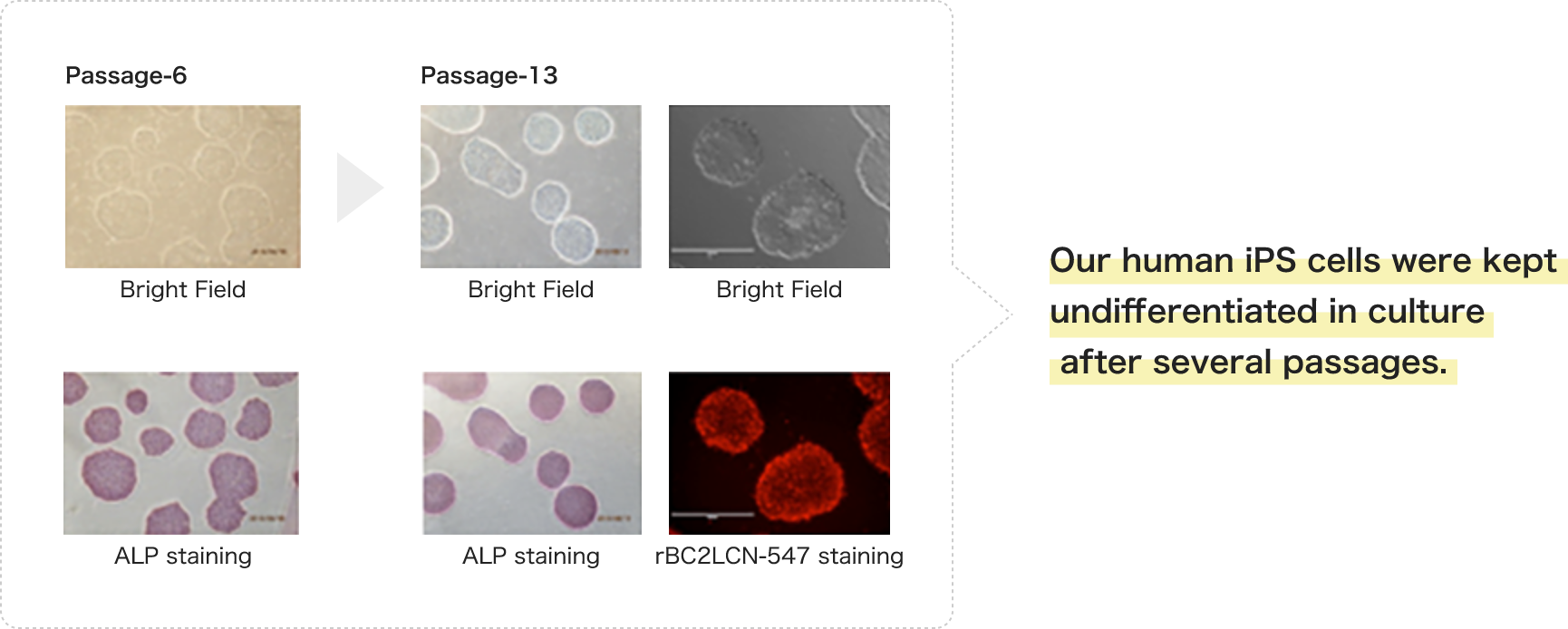
- Alkaline Phosphatase (ALP) Staining
Human Cell Reprogramming
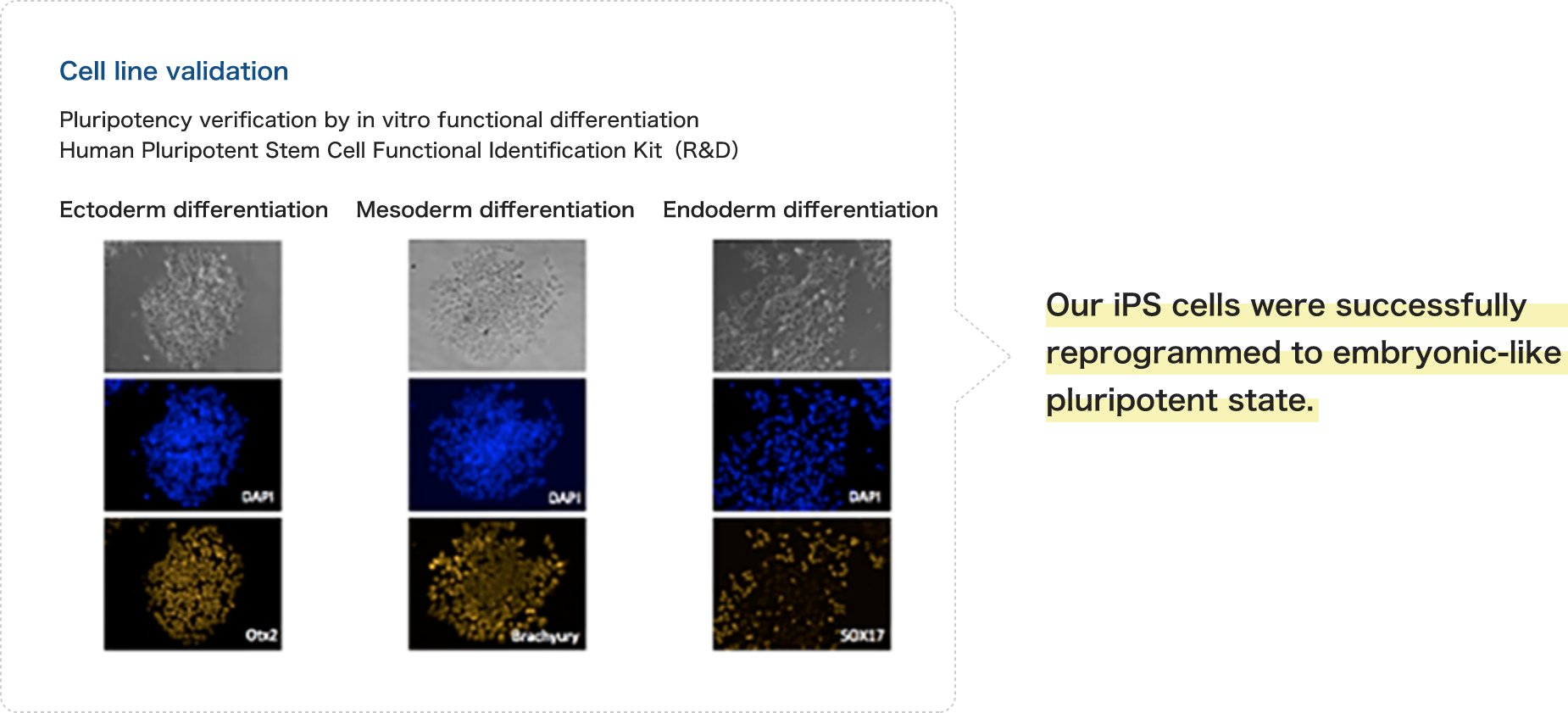
Cell line validation
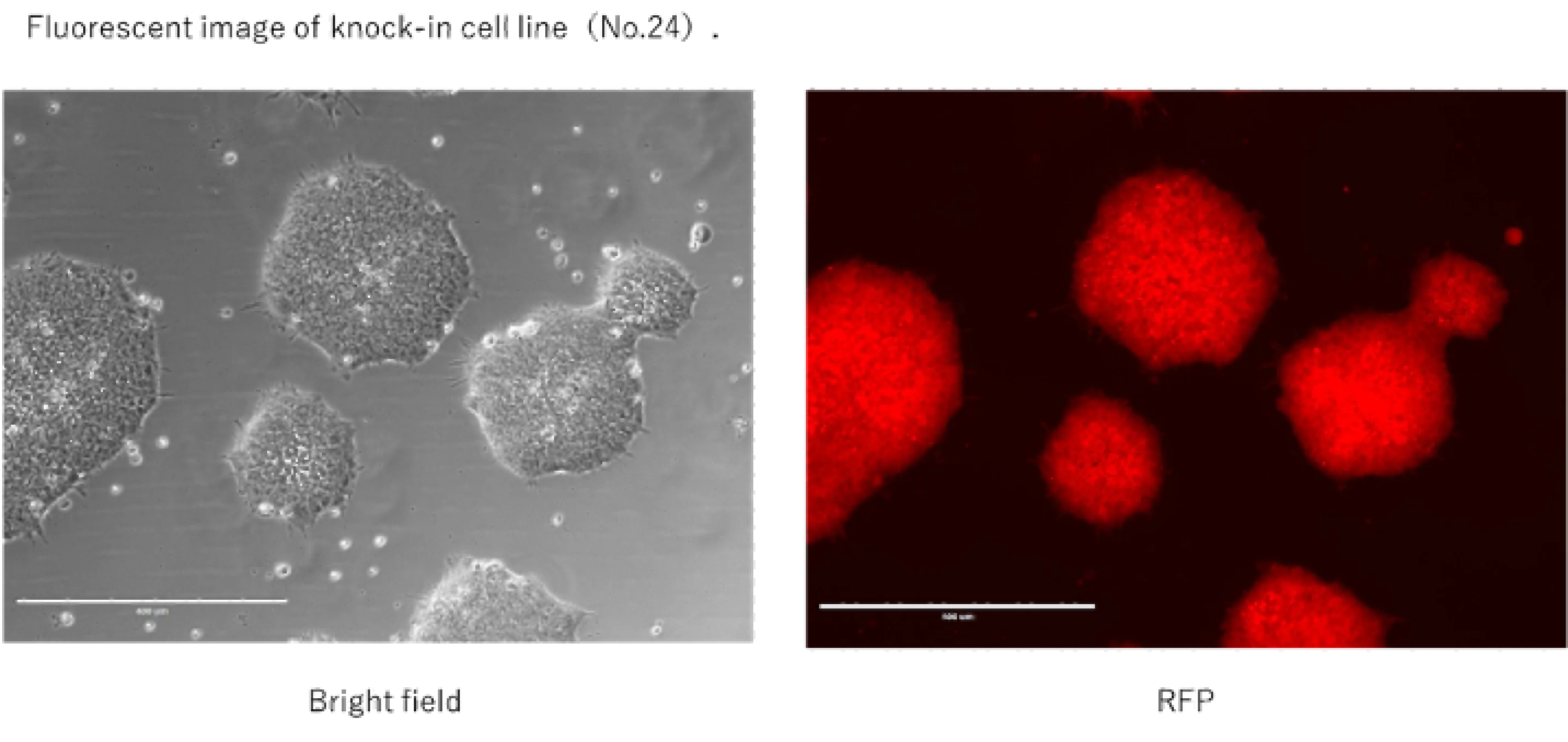
Genome editing of human iPS cells
Our contract service uses CRISPR/Cas9 to disrupt specific genes in human-derived cells that have been reprogrammed (iPS cells). In addition to gene disruption (frameshift/large deletion/stop codon knock-in), we can also perform point mutations and long-chain gene insertions using ssODN or plasmids as donors.
Contract generation of human iPS cell lines by genome-editting

Example for CRISPR -mediated targeted gene disruption of human iPS cell

Workflow for genome editing of iPS cells (estimated total work time: Knockout and short-chain knock-in: 4 months~. Long-chain knock-in: 6.5 months~).
-
Establishment of iPS cell line, culture, preparation of gRNA and donor DNA (1-4 months~).
We will thaw and expand the iPS cell lines※1,2 provided by the client, and then use them for genetic modification with the CRISPR/Cas9 system. The provided cell lines will be used after undergoing mycoplasma testing.
※1:We request that you provide two or more frozen vials from the same lot, with each vial containing at least 1×105 cells.
※2:For cell culture, we use StemFit® AK02N for feeder-free culture and STEM-CELLBANKER® for cryopreservation as our standard methods (other culture systems are available upon request).
We use “CRISPOR” (https://crispor.gi.ucsc.edu/) and “CRISPRdirect” (http://crispr.dbcls.jp/) to select a gRNA (crRNA + tracrRNA) sequence that is highly specific and common to both tools. We will also perform a sequence analysis of the candidate gRNA regions to confirm the sequence information.
When performing short-chain knock-ins of 1-10s of bases, such as point mutations, we design and prepare the ssODN that will serve as the donor DNA, in addition to designing the gRNA mentioned above. For long-chain knock-ins of several kilobases, we design and prepare the plasmid DNA that will serve as the donor DNA.
※3:The time required for Step 1 is approximately 1 month or more for knockout and short-chain knock-ins, and 4 months or more for long-chain knock-ins (this may vary depending on the length of the synthesized DNA, the complexity of the base sequence, and the difficulty of construction).
※4:For knock-in, the key to success is whether a gRNA can be designed near the desired mutation site. Therefore, we may not be able to accept cases where the design is difficult. Additionally, for short-chain knock-ins, in addition to the desired mutation, we also introduce silent mutations necessary to prevent re-cleavage by CRISPR and for genotyping (PCR-RFLP with the addition of a restriction enzyme site). -
KI by electroporation and selection of candidate cell populations (bulk cells) (approx. 1 month~).
We will prepare a complex of gRNA and Cas9 protein (RNP) targeting the gene of interest (plus donor DNA for knock-in) and introduce it into iPS cells using the 4D-Nucleofector™ (LONZA).
After culturing the candidate cell population (bulk cells) for a certain period, we will extract the genome and perform genotyping (PCR, etc.) to verify whether the expected mutation is included.
※5:For long-chain knock-ins, we perform drug selection using the drug resistance gene incorporated into the donor DNA. -
Clone selection and genotyping by single-cell cloning (approx. 1-2 months~).
Based on the bulk cells that showed the presence of the expected mutation in step 2, we will perform clone selection by single-cell cloning.
For clones derived from a single cell, we will perform genotyping (PCR, etc.) to confirm the presence or absence of the mutation (positive/negative). -
Vial stock of positive candidate clones and confirmation of positive clones by sequence analysis (approx. 1-1.5 months~).
For the positive candidate clones obtained in step 3, we will perform vial stock and sequence analysis on a subset of the clones to confirm the positive clones.
※6:We will prepare two vials of stock, with each vial containing at least 1×105 cells.
※7:If a large number of candidate clones are obtained, we will narrow down the analysis targets in consultation with the client.

